If you have any question, please contact:
Cuixiang Lin (lincxcsu@csu.edu.cn) or Jianxin Wang (jxwang@mail.csu.edu.cn).
The usage for the three functionalities are detailed separately below:
1. Singe gene analysis
2. Gene set analysis
1. Single gene analysis
Heterogeneous molecular networks (including PPI, coexpression, AD-associated miRNAs-target, sequence similarity and transcriptional regulatory networks) and AD-related trait data (including CERAD, Braak and CDR) are utilized to investigate and visualize the assocation of individual genes with AD. Following the two steps below, you can obtain and visualize how the queried gene is associated with AD based on the various networks and AD trait data.
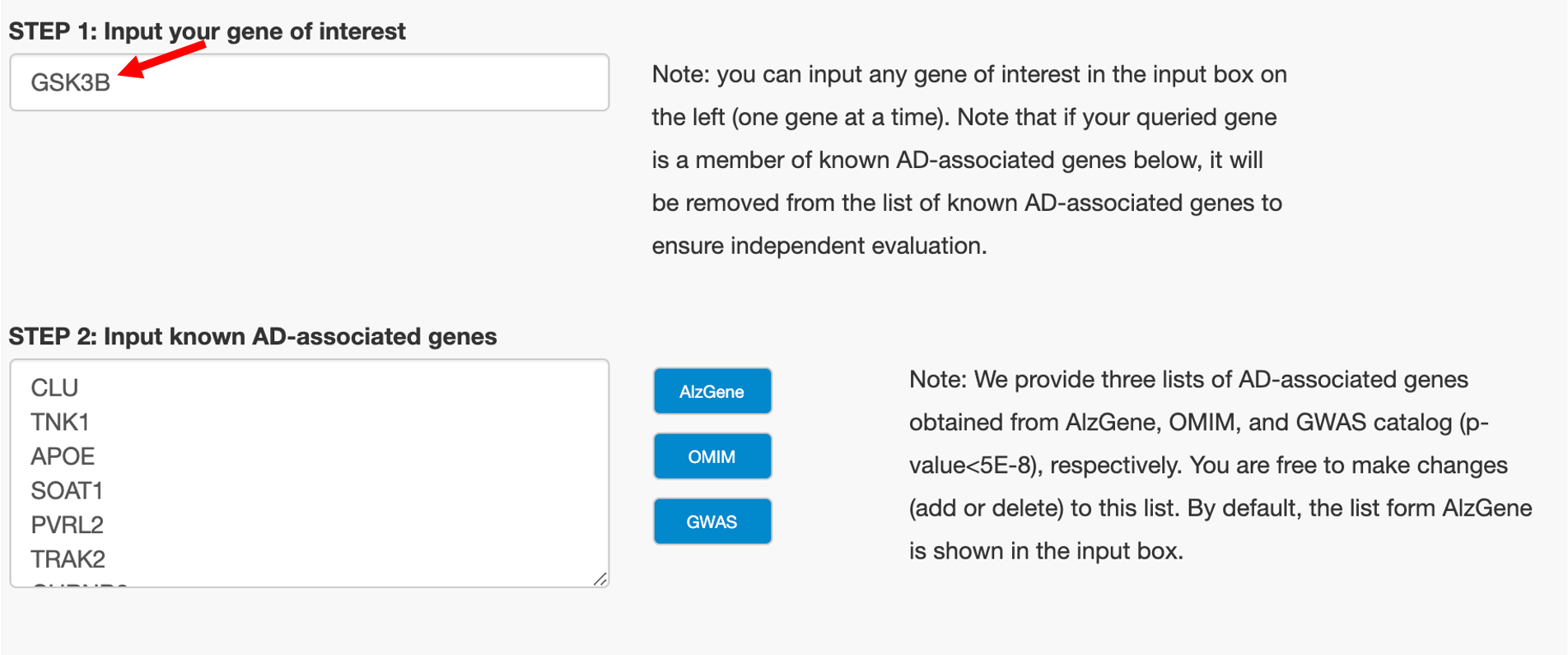
Step1: Input Your Gene of interest
Note: you can input any gene of interest in the input box on the left (one gene at a time). Note that if your queried gene is a member of known AD-associated genes below, it will be removed from the list of known AD-associated genes to ensure independent evaluation.
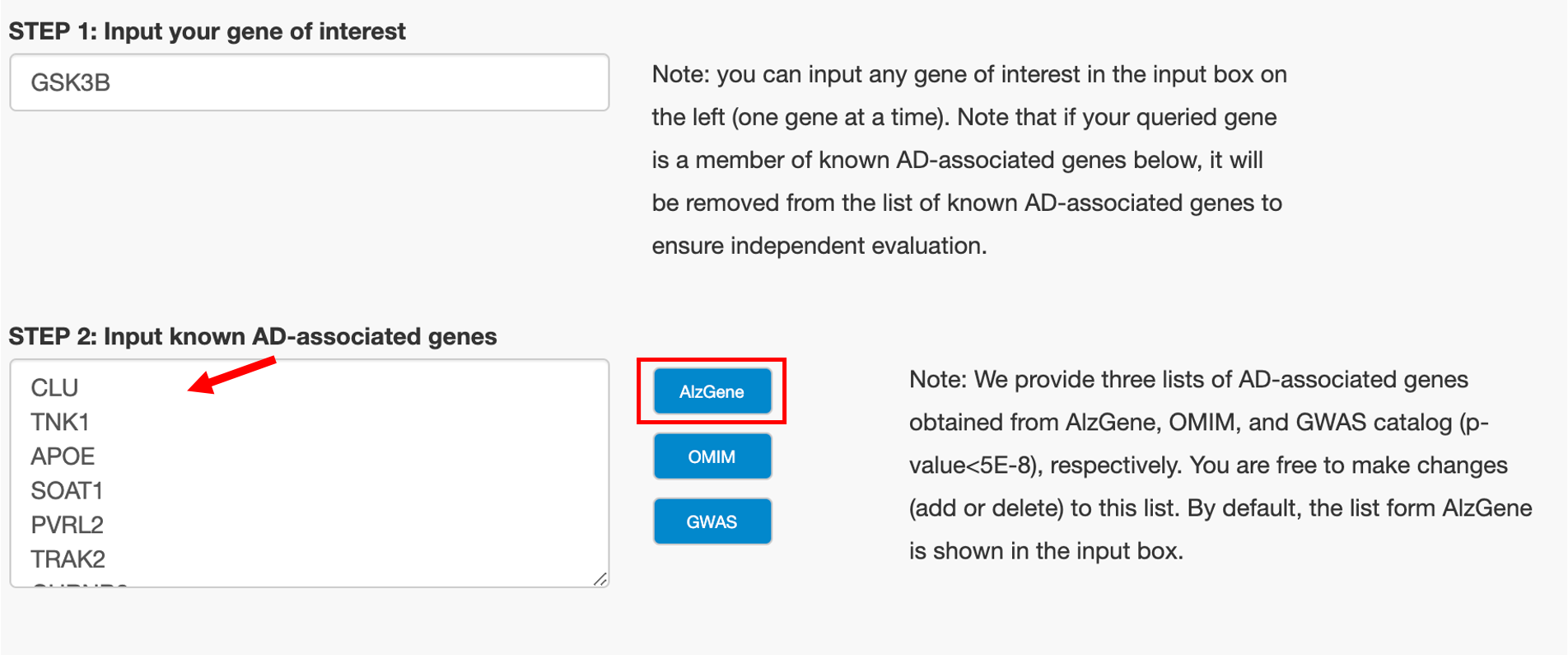
STEP 2: Input Known AD-associated Genes
Note: by default, AD-associated genes from AlzGene, OMIM and GWAS are provided. However, you are free to make changes (add or delete) to this list.
Notes:
results of cell type
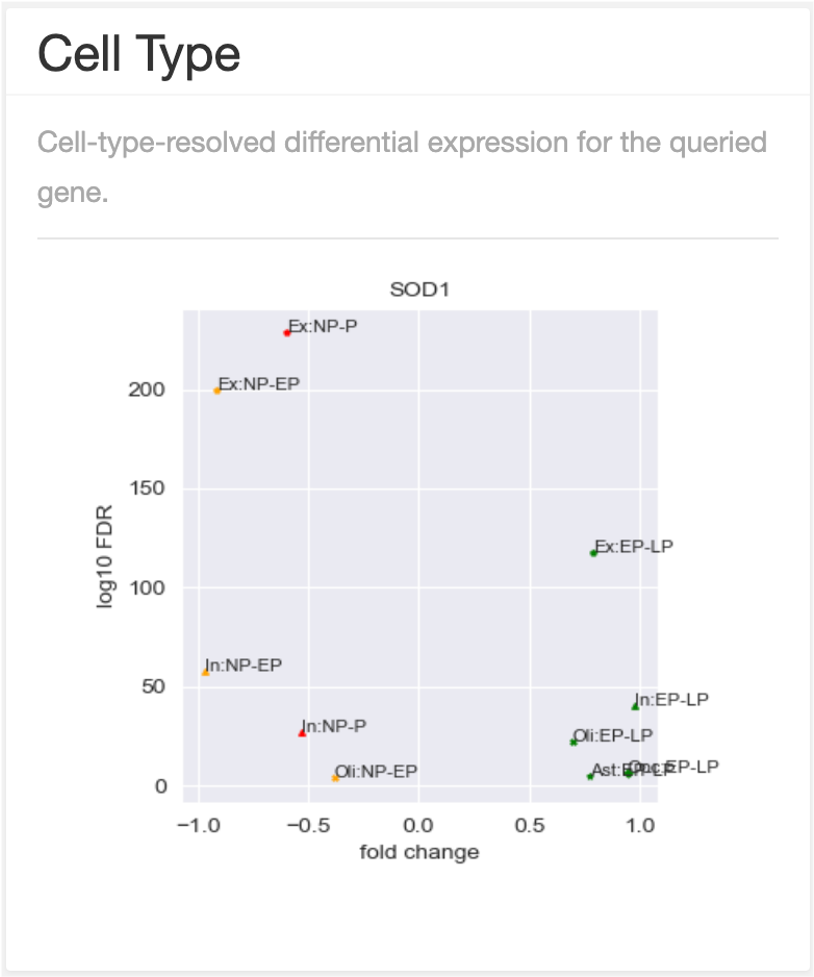
The cell-type data is from a study of Single-cell transcriptomic analysis of AD. It covers three stages and six major brain cell types. The stages includes no-pathology (NP), early-pathology (EP) and late-pathology (LP). The brain cell types are excitatory neurons (Ex), inhibitory neurons (In), astrocytes (Ast), oligodendrocytes (Oli), microglia (Mic), and oligodendrocyte progenitor cells (OPC). The annotations on figure are combinations of cell types and stages of AD. For example, 'Ex: NP-P' means no-pathology vs. pathology (both early pathology and late pathology) in excitatory neurons.
2. Gene set analysis
Geneset analysis evaluates the functional relationship between a gene set of your interest and the set of known AD-associated genes by interrogating heterogeneous biological networks. Please follow the instructions.
STEP 1: Input genes of your interest
Three example gene sets are provided here. The first gene set includes 50 genes from AD-associated biological process which is response to amyloid-beta (GO:1904645). The second one contains 28 genes which is about AD-associated phosphorylation event. The third one includes 50 genes which are least associated with AD from a study about AD. You are free to make changes to the gene set. Note that if any gene in the list is a member of the list of AD-associated genes that is used in Step 2, this gene will be removed from the list of AD-associated genes.
STEP 2: Input Known AD-associated Genes
There are 68 AD-associated genes from AlzGene showing as default AD-associated genes. The other two resources are from OMIM and GWAS. However, you are free to make changes (add or delete) to this list.
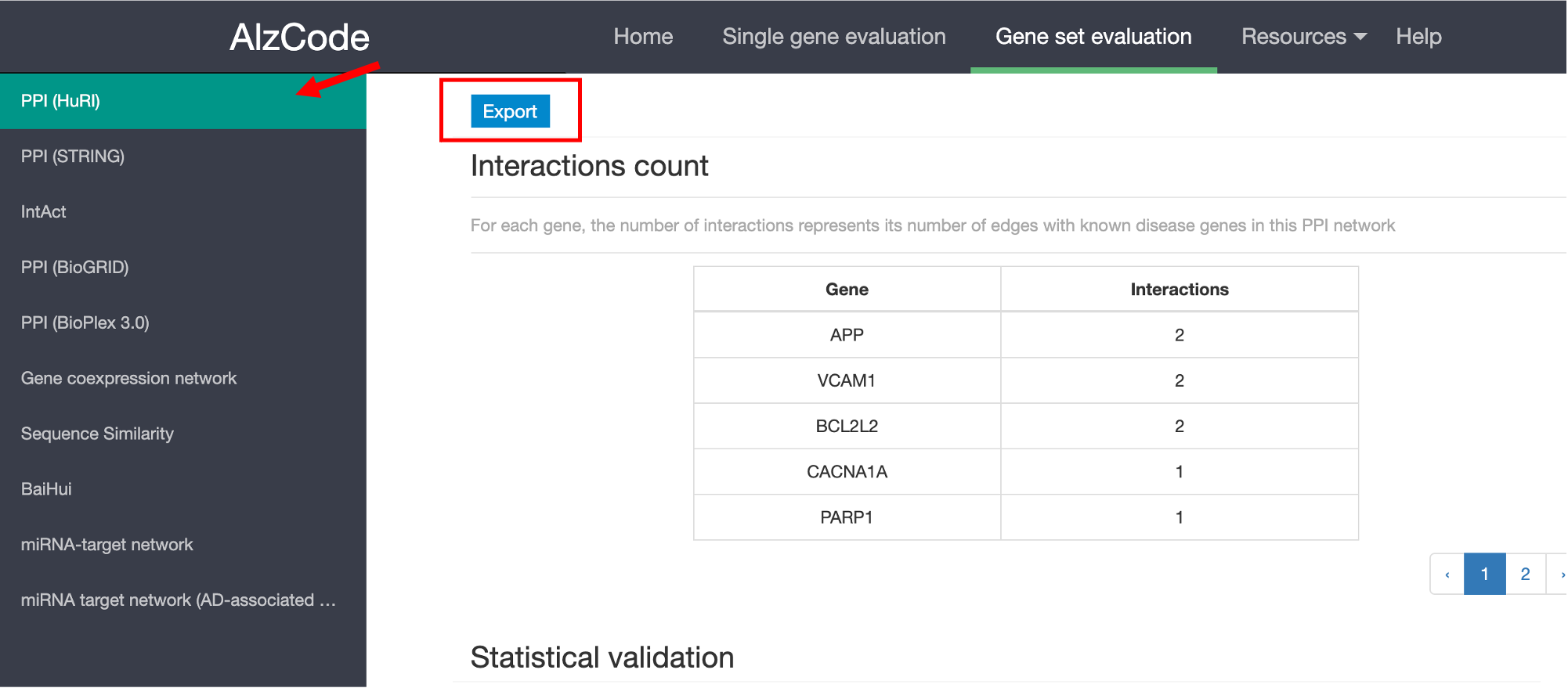
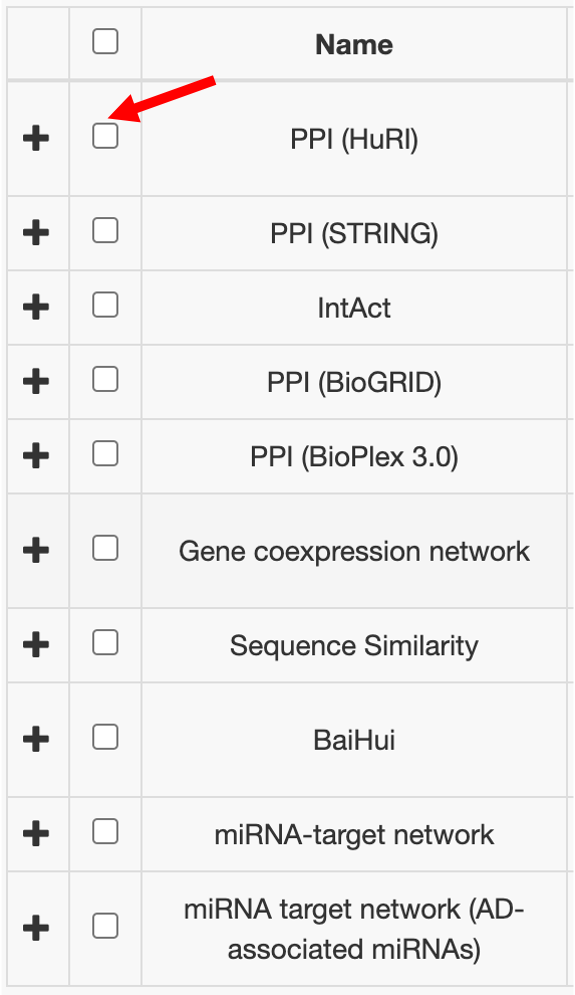
STEP 3: Select Molecular Interaction Networks (you can choose one or multiple networks simultaneously)
The networks are used to evaluate how significant the queried gene set interacts with the list of AD-associated genes compared to random baseline. The significance is measured with a p-value calculated by permuation test.
Submit
After completing the above three steps and clicking the 'submit' button, you will be directed to the result page as shown below. You can click each item on the left to view the results corresponding to each validation. A link is provided to download the interaction data with known disease genes for the queried gene set, statistical validation and subnetworks between queried genes and known disease genes.