AlzCode is a web resource that developed pipeline to evaluate the association of genes with AD. The association of genes with AD are validated by multiple lines of functional genomic evidence with statistical approaches. This web interface provides the following functionalities below:
Single gene evaluation
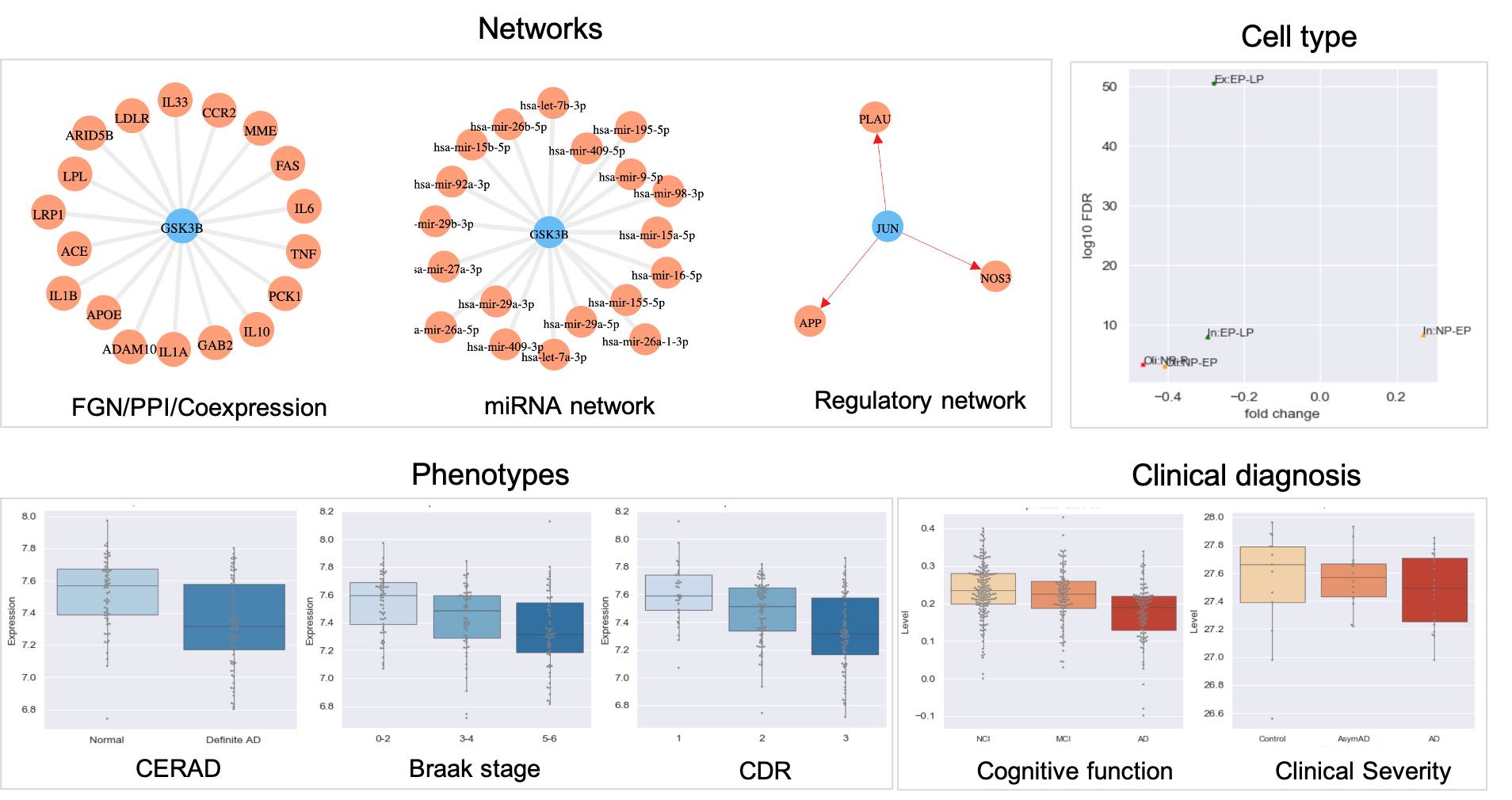
Single gene evaluation : Exploit and visualize the association of an individual gene with AD through both heterogeneous molecular interaction networks including brain-specific functional network, protein-protein network, coexpression network, miRNA network, sequence similarity network and regulatory network, AD-associated cell type information, AD-related traits (including CERAD, Braak and CDR) and clinical diagnosis (cognitive function and clinical severity).
Gene set evaluation
Gene set evaluation: Provide access to statistical approaches to evaluate the association of queried gene sets with AD by interrogating heterogeneous networks such as PPI, coexpression and miRNA-target binding networks.
Citation: Cui-Xiang Lin, Hong-Dong Li, Chao Deng, Shannon Erhardt, Jun Wang, Xiaoqing Peng, Jianxin Wang, AlzCode: a platform for multiview analysis of genes related to Alzheimer’s disease, Bioinformatics, Volume 38, Issue 7, 1 April 2022, Pages 2030–2032, https://doi.org/10.1093/bioinformatics/btac033
Copyright @ Hunan Provincial Key Lab on Bioinformatics
School of Computer Science and Engineering, Central South University, Changsha, Hunan Province, P.R. China